
Written By: Biomeme Staff
Currently, vector-borne diseases account for more than 17% of all infectious diseases, causing more than 700,000 deaths annually. With increased global travel and urbanization, vector-borne disease outbreaks are expected to rise, increasing these already high numbers. To combat this deadly trend, new methods for detection and prevention must be developed and implemented. The World Health Organization (WHO) has developed a special tool for determining which diseases to prioritize, known as the WHO Global Priority Pathogens List. This tool aims to identify vector-borne diseases that pose a public health risk because of their epidemic potential and for which there are no, or insufficient, countermeasures. A new list is anticipated to be released soon, but the latest list of priority diseases dates back to 2018 and includes:
These diseases are no joke. Zika can cause severe birth defects, CCHF outbreaks have a case fatality rate of up to 40%, and approximately 35% of reported patients with MERS-CoV infection have died. Unfortunately, because of the nature of how these diseases are spread it is difficult to prevent and control them. This is especially true because vaccines are available for only a few of these diseases.
Below you’ll find a brief overview of three relatively newer methods for detecting and fighting vector-borne diseases using tick and mosquito species as examples. Implementing even one of these approaches could positively and immediately impact biosurveillance efforts. If you’re interested more specifically in how Biomeme’s latest technology helps ongoing biosurveillance missions, head over to our Vector Surveillance page for more information.
Before genetic speciation, the identification of species such as ticks was traditionally done using morphological characters (i.e. the physical structure of the tick). In fact, this method is still in use today, but as you can imagine, organisms, especially tick nymphs (which can be the size of a pinhead), are often challenging to speciate based solely on their shape, size, color and other structural features. Take for example this tick nymph attached to our co-founder, Jesse vanWestrienen’s, thigh.
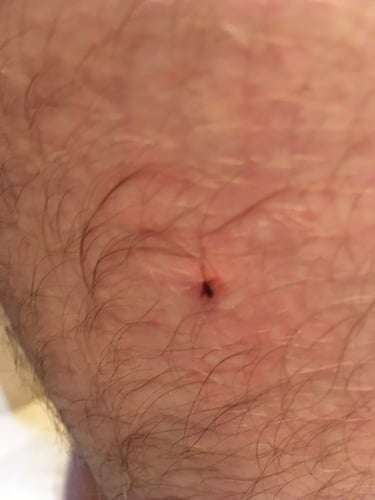
It was a Lone Star tick and fortunately for our head of biology, no pathogens were detected.
Speciation of ticks, like the one embedded in Jesse, helps to better identify potential pathogens as specific pathogens are associated with particular species of ticks and not others. Speciation can also inform the design of vector management programs - for example, certain mosquito species are active at night versus during the day.
Genetic Speciation enters the chat.
New rapid real-time PCR assays like our Tick + Pathogen Test targets specific DNA genes not only for the detection of pathogens, but also to genetically speciate amongst the most commonly occurring north american ticks: Ixodes scapularis (Deer Tick), Dermacentor variabilis (American Dog Tick), and Amblyomma americanum (Lone Star Tick). There are several methods by which we can specifically identify species using qPCR.
The most straightforward of these approaches is to target a gene or region of a gene that is unique to the species that you want to identify. This approach works very well to identify one species from another and is used in our Tick + Pathogen Test. However, if you want to separate two closely related species, or subspecies with very similar genetics a different approach is required.
Single nucleotide polymorphisms (SNPs) detection is a common approach used to genetically separate closely related species or detect specific mutations of importance. DNA is made up of nucleotides (Adenine (A), Thymine (T), Guanine (G), and Cytosine (C)) and SNPs are mutations of a single base pair of DNA that occur at varying frequencies within the population. The detection of multiple SNPs can be used to genetically identify one person from another, determine ancestry, or solve crimes in a process known as DNA fingerprinting. In the realm of vector surveillance, SNPs can be used to separate species or more often to detect specific mutations that confer resistance to commonly used pesticides. The rapid detection of resistance causing mutations allows for the timely and effective use of insecticides decreasing the prevalence of vector borne disease.
A more complicated approach would be to amplify a section of DNA common to several species that contains what is known as a barcoding region such as the 16s rRNA gene. Amplification of this region can be followed by sequencing and analysis of the barcoding region. While this approach is more complicated and requires sequencing in addition to PCR, it is very powerful and allows for the discovery of novel emergent disease vectors.
As a result of these methods, the adoption of genetic speciation via real-time PCR has helped overcome the cumbersome and sometimes unreliable morphological identification of closely related tick species in regions all over the world.

Byard (2016) reports that “every year, millions of people are bitten by the mosquito Aedes aegypti and thousands die as a result of infection by the viruses the mosquito carries”. To combat this, pesticides are typically used in an attempt to control the mosquito population, but mosquitoes swiftly develop resistance to these chemicals. Appropriately, biosurveillance teams have responded using molecular biology to overcome pesticide resistance by:
Two common mosquito pesticide resistance mutations that can be detected via real-time PCR are:
While a pesticide-free world is still a long way away, scientific breakthroughs such as this go a long way in targeting and ultimately disrupting the population of mosquitos and the sometimes deadly vector-borne diseases they carry. If mosquito surveillance is of particular interest to you, consider checking out our Mosquito-borne Pathogen Panel here.
It’s challenging to predict and control vector-borne disease outbreaks without some sort of geographical analysis, like surveillance maps. With reliable data, digital surveillance maps enable biosurveillance teams to more accurately identify and target areas with high indices of mosquito and tick infestations.
By leveraging API web services, such as our Web API, you can consume vast amounts of critical data faster and simpler than ever before. It requires engineering resources, but with something as simple as a tick or mosquito map, you can better target your efforts to high risk areas in order to eliminate the likely origin of tick and/or mosquito vectors.
It’s early in a new decade, and many R&D teams have goals to improve upon their tools to meet the most urgent missions directly at the point of need. Why not consider improving upon your team’s technical ability?
We recommend checking out VectorMap by Walter Reed Biosystematics Unit. It’s a unique resource for collection records, distribution models, and geospatial data of arthropods of biomedical importance, designed to inform risk assessments of vector-borne diseases.
The Navy Entomology Center of Excellence (NECE) is the Navy and Marine Corps center of expertise for operational entomology. They develop and evaluate novel products and application technologies to better protect deployed forces from blood feeding insects and other arthropods that transmit human diseases. NECE’s Project Sea Raven is focused on providing a highly mobile, complete pathogen surveillance and warfighter protection kit that can be deployed anywhere in the world with their Sailors and Marines.
We’ve developed a series of rapid mobile sample extraction protocols and shelf-stable real-time qPCR assays for use on our Franklin system to enable field-based sampling, genetic speciation and molecular detection of mosquito- and tick-borne pathogens for NECE’s Project Sea Raven.
A more detailed case study is coming soon so stay tuned!
In the meantime, here's what's on deck for the Biomeme/Project Sea Raven collaboration:
Subscribe to our monthly newsletter to get notified about the upcoming Project Sea Raven Case Study:
Get the latest tips from Biomeme shipped right to your inbox

Host response testing allows physicians to quickly rule out entire categories of potential infection to narrow their focus to the likely culprits for an illness. This can enable physicians to make...
401 North Broad St Suite 222 Philadelphia, PA 19108